About Me
I’m a graduate student at the University of Notre Dame in the Integrated Biomedical Sciences PhD program. I’m a member of two labs in the Chemistry and Biochemistry Department: the Champion Lab and the Whelan Lab. I am also a CBBI Fellow, a Berry Family Fellow at the Berthiaume Institute for Precision Health, and a Lucy Family Institute for Data and Society Graduate Scholar. My projects focus on method development in bottom-up proteomics, both in sample preparation as well as data analysis and informatics. I use capillary electrophoresis, liquid chromatographay, and mass spectrometry in the lab, and I develop accessible informatics tools using R and python for the analysis of resulting proteomics data.
Prior to graduate school, I worked as an associate scientist at Pharmaceutical Product Development (PPD) in Biopharmaceutical Method Development & Validation.
Research
Champion Lab - Small protein detection and characterization
Small proteins (S-prots) are an understudied area of the proteome, and are often missed in normal proteomic workflows due to loss in sample preparation as well the lack of genetic information which is required for database searching. I am developing tools to identify these proteins using Ribo-seq data and Data Independent Acquisition (DIA) along with informatics tools to analyze the resulting spectra. Additionally, I have developed a web app called PrIntMap-R which has a suite of tools available for protein coverage visualization and comparison of shotgun proteomics datasets. This tool won the 2023 Notre Dame Center for Research Computing Award for Computational Sciences and Visualization.
Whelan Lab - Ovarian cancer biomarker characterization
MUC16 is a large, heavily glycosylated mucin protein that is a biomarker used to track progression of ovarian cancer. The Whelan lab has made significant improvements on the peptide coverage of MUC16 utilizing a bottom-up proteomics approach. I am working to identify additional glycosylated peptides that can be used to not only characterize MUC16 expression in cancer patients, but also differential glycosylation. We believe that this will lead to new methods to detect ovarian cancer and ovarian cancer recurrance. I recently published a paper detailing how I have used preparative capillary electrophoresis (CE) to improve the proteomic coverage of MUC16, and how the technique may be applied to the characterization and detection of biomarkers.
Teaching
In addition to TA experience in Biochemistry, Organic Chemistry, and Cell Biology lab classes, I have developed and taught a programming activity that is designed for undergraduate chemsitry students. This freely availible resource guides novice programmers through using and writing code to analyze data that is relevent to analytical chemistry students. Through survey data from students who have done the activity as part of their class, I have found that the assignment is enjoyable, effective, easy to use, and students said they are likely to use the learned skills in their own research.
Photos and Figures
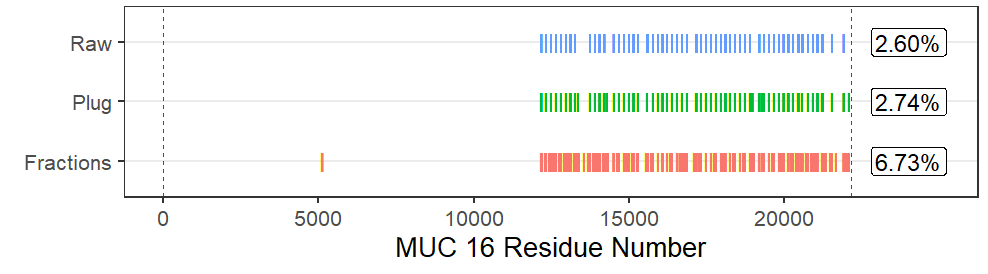 Increased MUC16 peptide coverage using CE Fractionation. Identified peptides shown with vertical colored lines.
Increased MUC16 peptide coverage using CE Fractionation. Identified peptides shown with vertical colored lines.
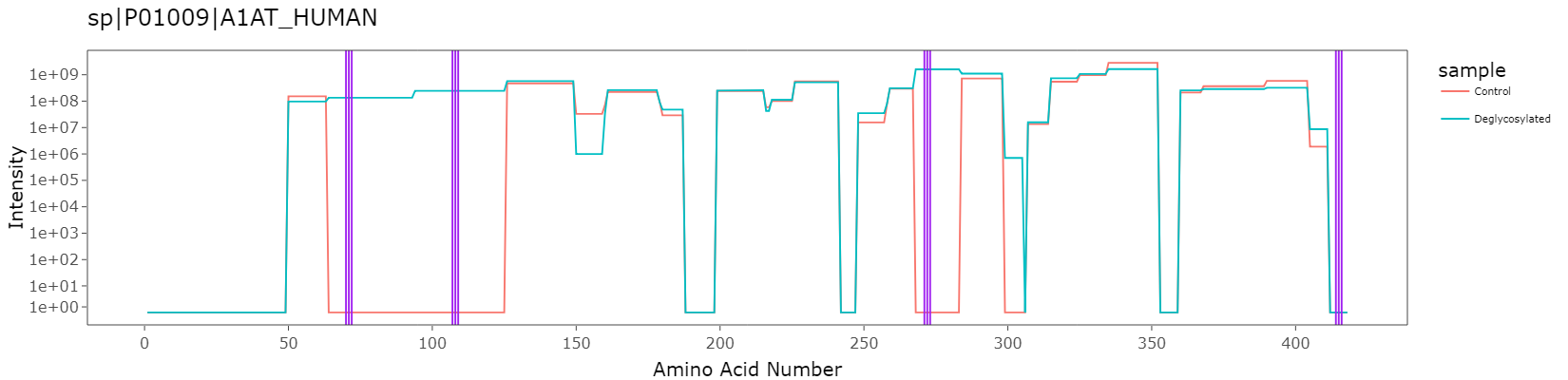 Intensity vs Amino Acid position for protein P01009 for a control sample and a enzymatically deglycosylated sample. Potential N-glyco sites highlighted in purple. Export from PrIntMap-R
Intensity vs Amino Acid position for protein P01009 for a control sample and a enzymatically deglycosylated sample. Potential N-glyco sites highlighted in purple. Export from PrIntMap-R
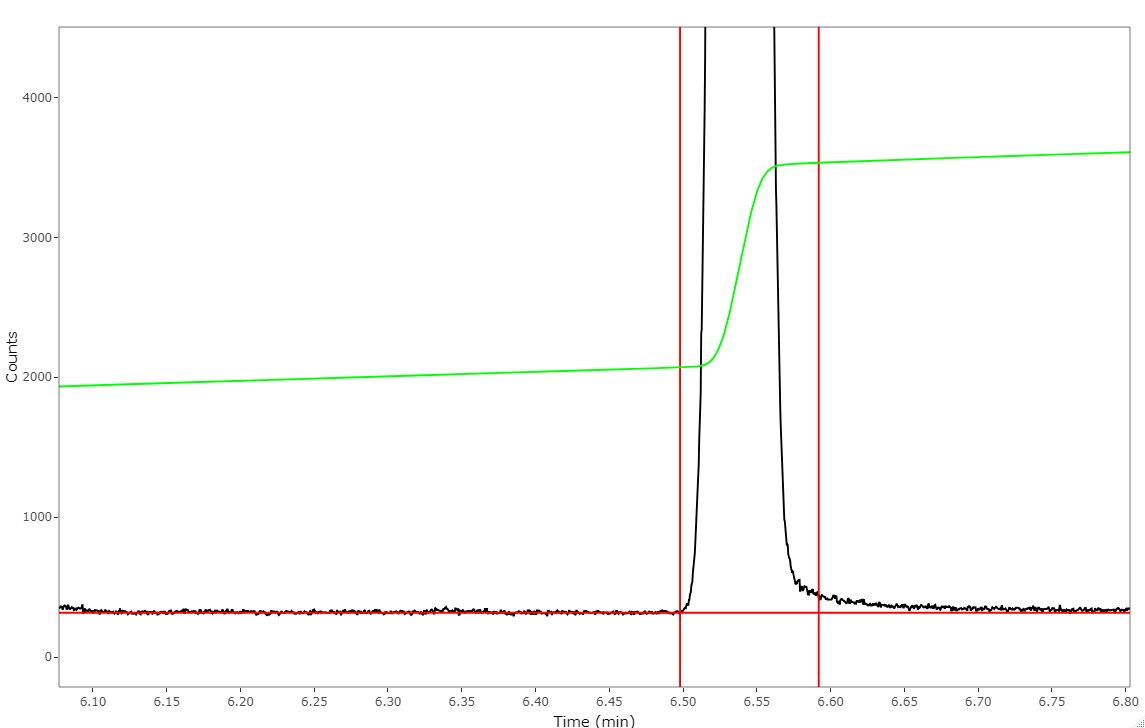 R Script to plot and integrate the raw data collected by the Avalanch Photodiode (APD) detector from the home built CE-LIF system.
R Script to plot and integrate the raw data collected by the Avalanch Photodiode (APD) detector from the home built CE-LIF system.
